Hayley Thompson has finally found her ‘ideal job’ – identifying pathogens in rodents – here at NRI, University of Greenwich. Her career journey to become a Molecular Microbiologist has taken her on a circuitous route, and has involved a variety of roles and projects, juggling work with childcare and long commutes along the way. Hayley takes up the story:
 When I left school I worked in several different office-based jobs, but never really enjoyed them. Inspired by my A-level biology teacher, who talked in great detail about her career in research, I decided to do a degree in molecular biology here at the University of Greenwich at the Medway campus.
When I left school I worked in several different office-based jobs, but never really enjoyed them. Inspired by my A-level biology teacher, who talked in great detail about her career in research, I decided to do a degree in molecular biology here at the University of Greenwich at the Medway campus.
After quite a varied career, I’ve now come full circle from studying here, to finding my ideal job as a molecular microbiologist at NRI. Working with Professor Steve Belmain - a world expert on rodents - and some of the team in the Agriculture, Health and Environment Department, is absolutely fascinating.
When I was studying for my BSc, the programme offered by Greenwich covered so many interesting elements of molecular biology – cancer biology, genetics, medical microbiology, bio-chemistry – it really was varied. I did a 4-year course, the third year of which was an industrial placement, which I spent in the laboratories of Pfizer. I really enjoyed the hands-on element of being in the lab and doing experiments and I decided a career in research was for me.
I was a mature student and was incredibly focused. I wasn’t that preoccupied with the social side of things, so I was able to put all my energy into my studies and I graduated with 1st class honours.
During exams, I became overcome with nerves, or adrenaline, and once I knew I was in line for a 1st, the pressure was really on, and it affected me hugely. I was able to deploy lots of calming techniques to get through.
On completion of my degree, I was accepted on to a PhD at King's College London in Microbiology. My research involved using a technique called IVIAT – In-vivo Induced Antigen Technology – to identify virulence factors in the bacterium Tannerella forsythia, a putative periodontal pathogen - bacteria that significantly contributes to periodontitis, a severe gum infection that can lead to tooth loss and other serious health complications.
I got married in the second year of my PhD and as we’d been together for about ten years, my husband was used to my studying. Following my PhD, one of the professors in the department asked me if I’d be interested in working with him on a short project, so I decided to stay at King’s.
I was then offered a five-year post-doc, researching the oral microbiome of humans (a community of microorganisms, predominantly bacteria, found within the mouth), with particular interest in the species present that are not or cannot be routinely cultivated (known as ‘uncultivated’ species). About nine months in, I discovered I was pregnant. During my maternity leave, my research group moved from King’s to the Blizzard Institute at the Queen Mary University of London (QMUL).
research group moved from King’s to the Blizzard Institute at the Queen Mary University of London (QMUL).
When I returned after having my daughter, I was offered the choice of staying at King’s or moving to where my group now was. I moved to QMUL, but the commute was about two hours each way and that was on a good day! I lasted about seven months and then sadly decided to leave, as juggling childcare on top of a busy job and a long commute was just too much. It was such a hard decision and I feared it might be ‘game over’ in terms of my career in a job I loved.
I think many working mums have faced similar choices. Some end up working effectively for nothing because of the high cost of childcare, just to keep their career open until their children start school. I was taking home very little once I’d paid my commuting and childcare costs.
I ended up taking five years out just to enjoy my daughter. For a while it was incredibly hard for me as I felt I’d somehow lost my identity. But I adjusted, ended up enjoying the time off and had faith that when the time was right, I’d pick up my career again.
One day, my former boss contacted me out of the blue and said that he was back at King’s, had funding, and would I like to come back and work for him either full or part-time? I jumped at the chance and ended up working 26 hours over three days which was perfect. A degree of flexibility is absolutely essential for getting mums back into the workplace.
I went straight back into the project where I had left off – obviously lots of things had advanced in my absence, but I was able to continue my research on investigating microbial dark matter – the not-yet-cultivated bacteria that we are currently unable to grow in the laboratory. In collaboration with the Forsythe Institute in Boston, USA, as part of the Human Oral Microbiome Database project, I was developing techniques to grow these uncultivated bacteria in the lab, with a specific focus on the Candidate Phyla Radiation Bacteria, a huge group of bacteria that have been described as ultra-small due to their reduced size compared to other bacteria. They have reduced genomes and have been shown to be dependent on other bacteria to grow.
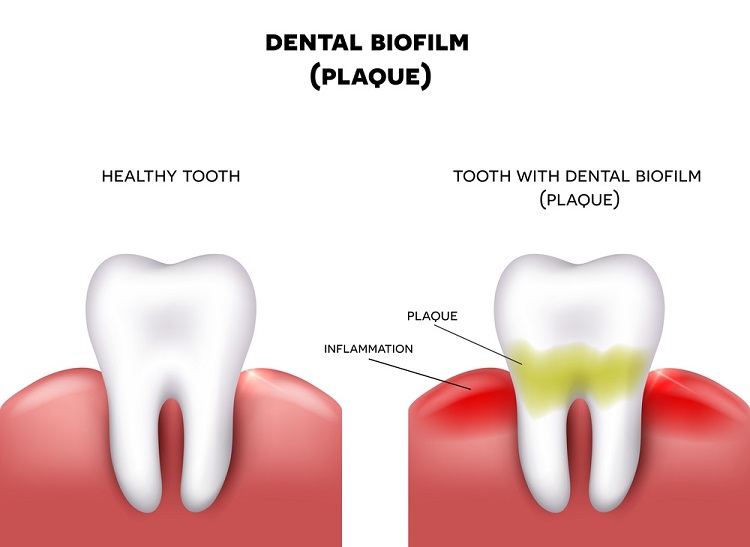 I managed to work for just a year before Covid came along and all work collecting saliva samples had to stop. At the end of the second year, I decided to grab another opportunity and switched to a new project with GSK (formerly GlaxoSmithKline plc, a British multinational pharmaceutical and biotechnology company). The project was focused on testing the efficacy of novel anti-microbials on oral biofilms – microbial communities inside the mouth that are normally known as dental plaque. My work on this project involved using little ‘pegs’ that mimic the tooth’s surface; oral biofilms are established on the pegs and then exposed to anti-microbials, after which the biofilms are removed, and the DNA extracted. The bacterial composition of the biofilms is then assessed using high-throughput sequencing – technology that can sequence multiple DNA targets in parallel, enabling millions of sequences to be generated at a time. This allowed me to develop my bioinformatics and sequencing skills further, something that I was eager to do.
I managed to work for just a year before Covid came along and all work collecting saliva samples had to stop. At the end of the second year, I decided to grab another opportunity and switched to a new project with GSK (formerly GlaxoSmithKline plc, a British multinational pharmaceutical and biotechnology company). The project was focused on testing the efficacy of novel anti-microbials on oral biofilms – microbial communities inside the mouth that are normally known as dental plaque. My work on this project involved using little ‘pegs’ that mimic the tooth’s surface; oral biofilms are established on the pegs and then exposed to anti-microbials, after which the biofilms are removed, and the DNA extracted. The bacterial composition of the biofilms is then assessed using high-throughput sequencing – technology that can sequence multiple DNA targets in parallel, enabling millions of sequences to be generated at a time. This allowed me to develop my bioinformatics and sequencing skills further, something that I was eager to do.
My new job here at NRI is to carry out all the molecular work on ‘RodentGate’, a project to detect the presence and prevalence of pathogens in rodents and their spread on farms to livestock, pigs and chickens in particular. Supported by NRI’s molecular biologists and virologists, Dr Sophie Bouvaine and Dr Gonçalo Silva, I’m identifying which pathogens these rodents might have, by extracting the DNA and RNA from various rodent tissues and looking for different pathogens, initially using molecular techniques including polymerase chain reaction (PCR) and qPCR, where the ‘q’ signifies ‘quantitative’, meaning that the technique is used to quantify a specific fragment of DNA.
It’s quite a targeted approach as we know what we want to look for, and then we apply a broader metagenomic approach - the study of genetic material recovered directly from environmental samples - to sequence everything that’s there and see if we uncover anything new or anything we didn’t think would be there.
The work is a mixture of ‘wet lab’ and ‘dry lab’. ‘Wet lab’, also known as ‘experimental lab’, refers to hands-on experimental work, performing various types of reactions in the lab which I love, whereas ‘dry lab’ allows me to use and develop my bioinformatic skills in the analysis of sequence data; it’s perfect and I really enjoy it.
The project is based in the UK and across the EU and we study samples from all over. It’s my first full-time role since having my daughter so there’s still a large element of juggling involved but even though I’m just a couple of months in, I think I’ve got a routine now!
NRI is a great place to work as there is so much going on and crucially, a lot of collaboration and information sharing. It’s unlike anywhere else that I’ve worked and I’m so glad to be back!
RodentGate is supported by the ERA-NET fund for International Coordination of Research on Infectious Animal Diseases; NRI’s funding comes directly through UKRI (BBSRC)
To find out more about

